Abstract
Background: Chronic GVHD (cGVHD) is the most significant cause of long-term morbidity and mortality after hematopoietic stem cell transplant (HCT). Improving patient outcomes requires a better understanding of biological mechanisms that underly disease, in order to develop novel risk-assignment biomarkers and therapeutic strategies. Naïve T cells (TN) are a prevailing risk factor for cGVHD, with mouse studies and clinical trial data demonstrating that graft depletion of TN cells decreases the frequency of cGVHD. Nevertheless, little is known about how TN cells progress to alloreactivity and, ultimately, to clinical cGVHD. In this study, we compared single-cell RNA sequencing (scRNA-Seq) from patients receiving unrelated donor transplantation (URD HCT) who developed mod-severe cGVHD, versus patients who were free from cGVHD, to identify disease-specific subpopulations of T cells that preceded disease.
Methods: This analysis was designed to identify immune profiles in blood at Day 100 that could distinguish patients who ultimately developed cGVHD from those who remained free of this disease. Blood was drawn from patients with hematologic malignancies undergoing 8/8 HLA-matched URD HCT after myeloablative conditioning, and who received CNI/MTX GVHD prophylaxis. We focused on Day 100 samples from patients with Gr 0-2 acute GVHD who did not require systemic steroids as our starting population, and then divided these patients into those who subsequently developed moderate-severe cGVHD ('cGVHD' group, n=8; median cGVHD onset = 181 days) and patients who never developed mod-severe cGVHD through at least 2 years post-HCT (termed 'operationally tolerant' patients, 'OpTol', n = 24). Flow cytometry of longitudinal samples was performed using 16 ten-color panels, characterizing T cell reconstitution, activation, exhaustion, and naïve/memory subpopulations. Bulk RNA sequencing was performed on flow-sorted CD4 T cells (within 3h of collection) at Day 100 on patients from two central lab sites (cGVHD = 7, OpTol = 9). Statistical modeling was performed using DESeq2, followed by differential expression and Gene Set Enrichment Analysis (GSEA) with clusterProfiler. scRNA-Seq was performed on flow-sorted CD4 T cells using the 10X platform (cGVHD = 7, OpTol = 5). scRNA-Seq data was batch corrected with Harmony, annotated with SignacX and analyzed with Seurat.
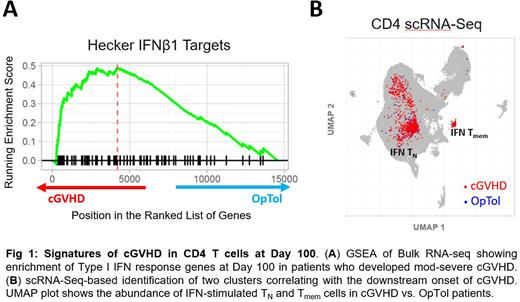
Results: Flow cytometry identified a higher proportion of CD4TN but not CD8TN cells in cGVHD patients that persisted longitudinally through Day 100 (25.9% vs. 10.0% at Day 100, p=0.03). Bulk transcriptome GSEA of Day 100 CD4 T cells confirmed enrichment of TN cells in cGVHD patients, but also revealed additional gene sets relating to interferon signaling (Fig. 1A). In contrast, OpTol patients revealed gene signatures relating to memory T cells and quiescence. scRNA-Seq enabled the discovery of CD4 T cell types/states enriched in patients who would develop mod-severe cGVHD. We profiled >100,000 CD4 T cells from Day 100 samples from cGVHD and OpTol patients. Unsupervised clustering identified two populations that were enriched in cGVHD vs. OpTol patients (Fig. 1B): interferon-stimulated CD4TN cells and interferon-stimulated CD4 memory cells. Both subsets show significantly higher expression of an interferon-response signature relative to all other cells (TN = 0.93, Tmem = 1.61, others = 0.14, p < 0.0001). The CD4TN subset possessed a signature for IFN-α-related stimulation (including IFI6, ISG15, IFI44L, EPSTI1) as well as IFN-γ-related genes (including STAT1, MX1). These findings further substantiate the IFN gene signature identified by bulk RNA-Seq at day 100 in cGVHD vs. OpTol patients.
Conclusions: These data identified an inflammatory CD4TN cell subpopulation at day 100 in patients that go on to develop cGVHD, and suggest that CD4TN cells may harbor pathogenic cells that could predispose patients to allo-inflammatory complications. scRNA-Seq revealed a strong type I and type II interferon signaling signature in this CD4TN subset, and also identified a novel population of interferon-stimulated memory cells which may indicate a more differentiated population of alloreactive cells. These data highlight an interferon-driven transcriptome signature preceding the onset of cGVHD. Continued exploration of these cells may yield new risk-assignment biomarkers and nominate therapeutic approaches to prevent cGVHD in high-risk patients.
Disclosures
Kean:EMD-Serono: Research Funding; Vertex: Consultancy; Mammoth Biosciences: Current equity holder in private company, Current holder of stock options in a privately-held company; Novartis: Research Funding; Bluebird Bio: Research Funding; Bristol Myers Squibb: Patents & Royalties: clinical trial, Research Funding; Magenta: Research Funding; HiFiBio: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal